Mutation Set | Sequence Activity Analysis | SARvision | Biologics
Using Mutation Sets to Identify Key Residues
by Conrad Y. Hansen
A mutation set shows Sequence Activity Relationships for a specific position.
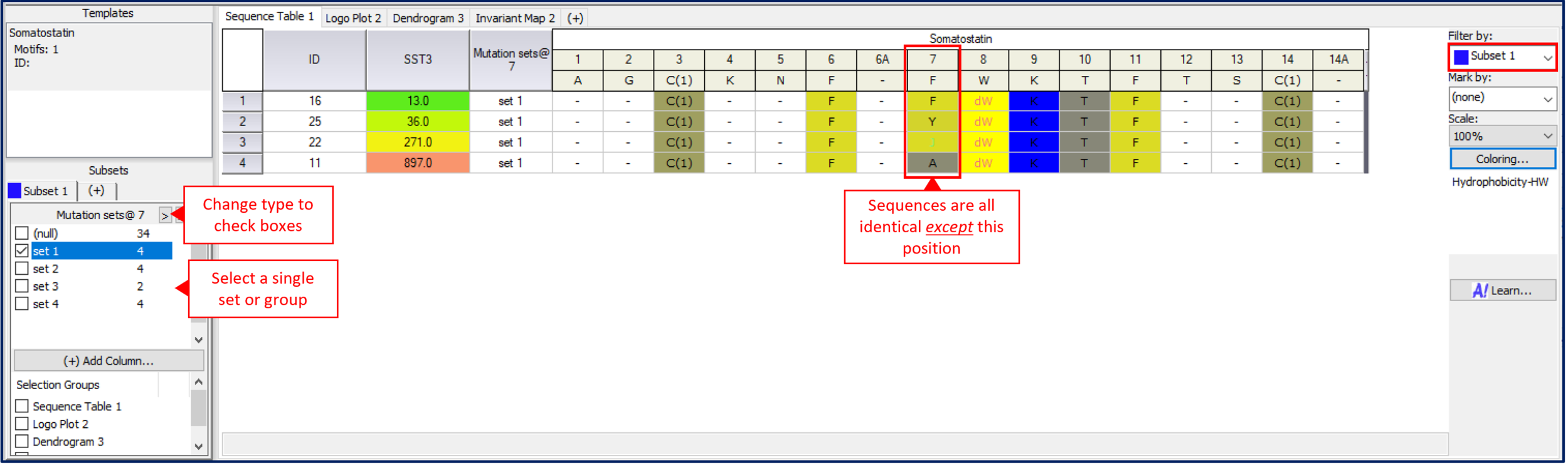
Mutation sets are similar to mutation cliffs; the key difference is that instead of identifying pairs of sequences that change at only a single position, mutation sets identify all sequences that are identical with the exception of a single position. A mutation set family forms a series where the substitution pattern at a single position can be directly correlated with changes in activity. These give confidence that the correlation is real and significant because all confounding mutations, or those located elsewhere in the sequence, are excluded in the analysis. Mutation sets are calculated for a single position at a time to create multiple sequence families where sequences are identical except at the chosen position. Under main menu->Tools->Calculate mutation set will build all possible sequence sets for a given position and adds it as a new column. Note that multiple mutation sets can exist for each position of the sequence creating a wealth of Sequence Activity Relationship data to explore.
Select a sequence alignment position to study, one at a time.
To analyzed the mutation sets interactively, add the new calculated mutation set column to the Subset filters (used the (+) Add column… in lower left). Change the filter to a text box and then select single mutation set families at a time (click the ‘>’ button). In the Sequence table, select filter by subset to show only this family. In this way, one can interactively navigate mutation set to identify trends in the data.
A mutation set is a family of sequences that vary at a single position, but are otherwise identical. Sequence Activity Relationships can be clearly observed in these sequence subsets.